Join us in congratulating Adeena on receiving the Provost's Undergrad Fellowship to study the 15q13.3 deletion haplotype using CiFi!! Her work will be presented at the annual Undergraduate Research, Scholarship and Creative Activities Conference this upcoming spring!
Ice Skating and Winter Party
The Dennis Lab came together for a winter social, starting with an ice skating outing and followed by a winter party. It was a great opportunity for lab members to relax, connect, and celebrate the season together.







2025 UC Davis Human Genomics Symposium
Gulhan Kaya and Nick Haghani presented at the School of Medicine (SOM) Human Genomics Symposium, sharing their ongoing research from the Dennis Lab. Nick presented work on the functional characterization of autism candidate genes associated with disproportionate megalencephaly using larval zebrafish, while Gulhan presented CiFi, a new low-input long-read method to unmask hidden chromatin. Together, the talks highlighted complementary experimental and computational approaches to studying human genomics and neurodevelopment.


UC Davis Chancellor's Fellow & Research Rock Star Basic Sciences Award
It’s been a good year for awards! Congrats to Megan and the whole team. Read more about Dennis lab research and the Chancellor’s fellowship here.
A huge congrats to the seven other awesome faculty that also received the Chancellor’s Fellowship (more info here) and the SOM Research Rockstars.
MIND Institute’s Dorothy Ross Graduate Student Research Award
Grad Student Nick Haghani has been awarded the Dorothy Ross Graduate Student Research Award for his proposal titled, “Functional Characterization of Autism with Disproportionate Megalencephaly Candidate Genes”. Congratulations!
The MIND Institute’s Dorothy Ross Graduate Student Research Award honors the memory of Dorothy Ross, mother of Dr. Len Abbeduto, and has been made possible through the generosity of Dr. Abbeduto and his wife, Terry McMenamin.
Gulhan receives inaugural Genome Center Award of Excellence in Science
So proud of Gulhan for her well-deserved award for contributions to many innovative genomic methods, including massively-parallel reporter assays, CiFi, sciRNA-seq, the list goes on-and-on!


The 2025 UC Davis Genome Center Halloween symposium was a success!
The Lego Legends of the Genome Center—otherwise known as the Dennis Lab—took home the award for Best Group Costume! Huge congratulations to Gulhan, whose incredible work on CiFi earned Best Poster, and to Nick for carving a very cool zebrafish–human–inspired pumpkin.





Hooray! Gabriana and Zoeb pass their QEs
Megan named to ASHG Board of Directors
We are celebrating this awesome news! Thank you to those that voted! Now, time to get to work.
Check out the ASHG press release here.
Welcoming Undergraduate Researchers!
We are excited to welcome undergraduate researchers:
Theo Gim, former 2025 FYS CURE student, who will be working with graduate student Zoeb Jamal to characterize the human duplicated gene NOMO1.
Olivia Mckenrick who will be working with graduate student Gabriana La exploring the impact of m6A mRNA modifications and processes on autism and fragile X syndrome using zebrafish
Ashley Ou, who will join our group as a UC Davis CIRM COMPASS fellow who will be working with postdoc Eleni Katsougia to characterize the functions of human duplicated (HSD) genes in zebrafish.
Shriya Kota, former 2025 FYS CURE student, who will also be working with postdoc Eleni Katsougia to characterize the functions of human duplicated (HSD) genes in zebrafish
David Martinez, who will be working with graduate student Nicholas Haghani to characterize the function of ASD associated gene CHD8 in zebrafish.
Small Samples, Big Insights: Capturing Chromatin Architecture with CiF
Ph.D Candidate Sean McGinty gives an On-Demand Webinar for Arima Genomics about CiFi. This webinar explores how the innovative CiFi method is advancing chromatin conformation capture, enabling detailed analysis of complex and repetitive genomic regions—even from low-input or challenging samples. This approach expands access to advanced 3D genomics by providing a robust workflow that overcomes limitations associated with sample size and complexity, allowing researchers to gain deeper insights into chromatin architecture across the genome.
Link: Small Samples, Big Insights: Capturing Chromatin Architecture with CiFi
2025 Human Genomics Focus Group Pilot Award
Congratulations to Gabriana La and Tanner Smith from the Baldrich Lab on receiving the 2025 Human Genomics Focus Group pilot grant! The award encourages creative interdisciplinary and cross-lab collaborations supporting findings that may translate into preliminary data for future grants and papers. Their proposed work for these funds aims to characterize how loss of m6A RNA readers impacts small RNA profiles and RNA modifications in zebrafish autism models. They will present updates on their findings to the Human Genomics Focus Group in Fall 2026.
International Zebrafish Conference (IZFC) 2025
Team Functional Genomics was excited to attend the 19th International Zebrafish Conference in Madison, Wisconsin 🧀 Five lab members presented their work:
Dr. Megan Dennis platform talk: “Zebrafish models of duplicated genes implicated in human brain evolution”
Grad student Nicholas Haghani platform talk: “Functional Characterization of Autism with Disproportionate Megalencephaly Candidate Genes in Larval Zebrafish”
Highlighted in this IZFC newssplash
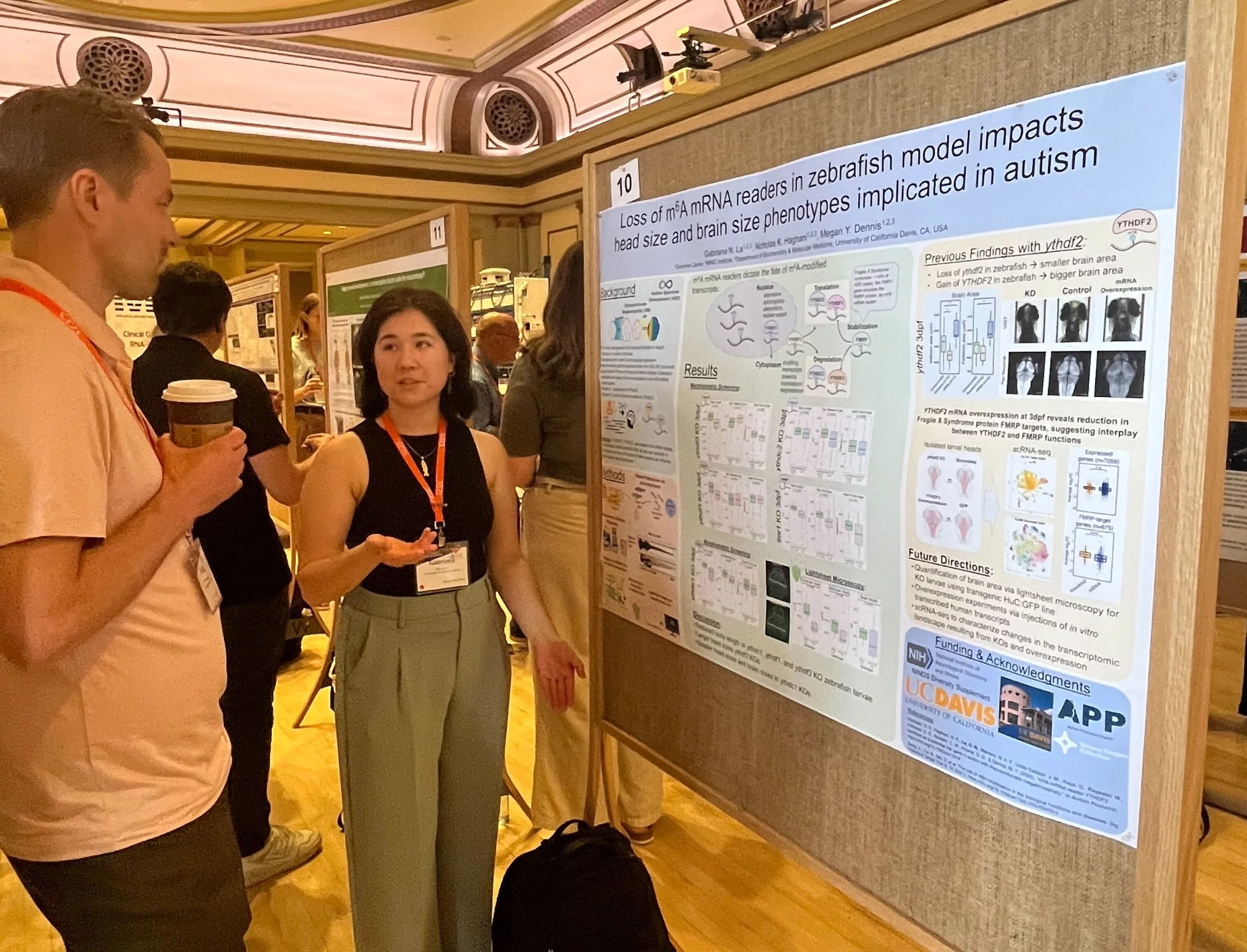
Grad student Gabriana La poster: “Loss of M6A mRNA readers in zebrafish model impacts head size and brain phenotypes implicated in autism”
Undergrad Aidan Baraban poster: “Characterizing brain functions of human duplicated NOMO paralogs using zebrafish”
Undergrad Emily Xu poster: “Characterization of an Autism Associated Gene KMT2E in Zebrafish”





Happy 10 Year Anniversary To Our Wonderful Lab!





Welcome to Daniela, our new junior specialist
Welcome (back) to Daniela Garcia, our new junior specialist and NIH PREP Scholar, who recently graduated with her B.S. in Wildlife, Fish and Conservation Biology from UC Davis!
Daniela will be working on our projects using zebrafish as a model organism to test genes implicated in neurodevelopment, and help maintain our zebrafish facilities.
Daniela also has an email address that any geneticist would be jealous of: dnagarcia@ucdavis.edu
Graduation picnic!
We have many graduations to celebrate!
Undergraduate research assistant Aidan Baraban graduated with a B.S. in Neurobiology, Physiology, and Behavior (and a minor in Music!). He is heading down to UC Irvine to pursue a PhD in neuroscience! Congratulations Aidan!
Undergraduate research assistant Emily Xu graduated with a B.S. in Molecular and Medical Microbiology with minors in public health and economics! She is currently working as an Integrative Medicine Assistant, volunteering at the Coalition of Concerned Medical Professionals, and studying for the MCAT. Good luck Emily!
Undergraduate research assistant Daniela Garcia graduated with a B.S. in Wildlife, Fish, and Conservation Biology! She will continue on in our lab as a Junior Specialist and NIH PREP Scholar. We are very lucky to have you Daniela!
Graduate student researcher Nabeel Sabzwari graduated with an M.S. in Computer Science and is currently interviewing for full time positions! In the meantime he is working remotely to wrap up a couple projects that he led in the lab.
Welcome to Eleni, our new postdoc
We are excited to welcome Eleni Katsougia, our new postdoc, who recently completed her PhD in Evolution and Genetics in the Ragsdale lab at Indiana University!
Eleni’s projects will focus on using zebrafish to learn more about the function of human duplicated genes.
Eleni also bakes the most amazing cakes!
Undergraduate Research Conference 2025
Our incredible Dennis Lab undergrads, Aidan, Emily, and Adeena proudly presented their work at the 2025 UC Davis Undergraduate Research Conference. Their posters highlighted months of dedication, curiosity, and scientific growth!
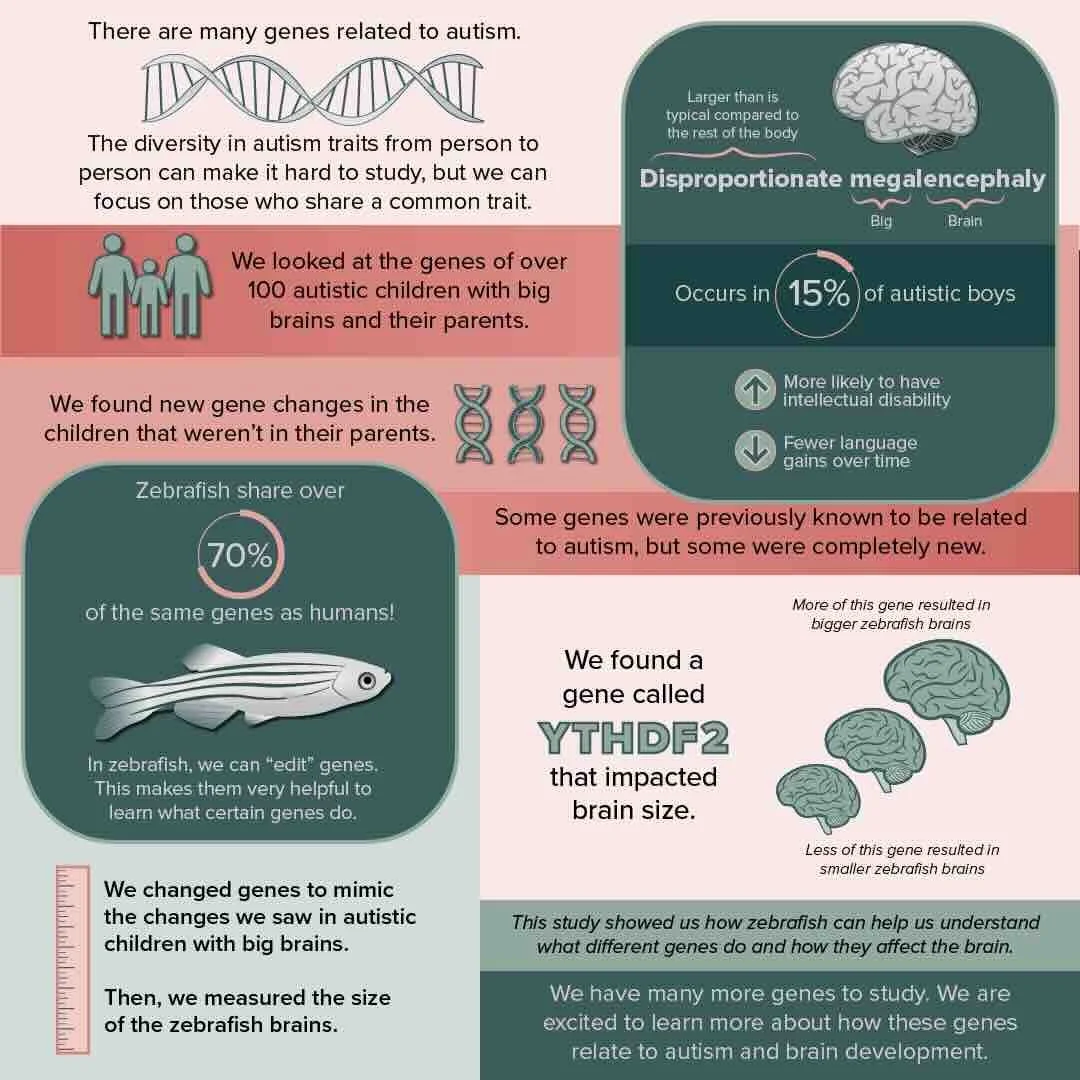
m6A-mRNA Reader YTHDF2 Identified as a Potential Risk Gene in Autism With Disproportionate Megalencephaly
We are excited to share our new paper published in Autism Research led by former postdoc Sierra Nishizaki, with support from many current (Nick, Gabriana, Gulhan) and former lab members (Tasha, José). Gabriana put together a great summary here.
In the study, we identified 154 candidate genes with likely gene-disrupting de novo or rare variants from whole-genome sequences of autistic children with disproportionate megalencephaly. We validated a patient duplication (gain of function) of m6A-mRNA reader YTHDF2 results in enlarged brain in a zebrafish larval model, while knockdown with CRISPR leads to microcephaly. Using single-cell RNA-seq of zebrafish larval brains points to possible downregulation of FMRP-associated genes with increased presence of YTHDF2. We also identified a loss-of-function variant impacting YTHDC1, implicating more broadly the m6A-modification pathway as possibly contributing to ASD.
The project would not have been possible without excellent collaborations through the UC Davis MIND Institute, including the Autism Phenome Project led by Drs. David Amaral and Christine Wu Nordahl, and genomic sequencing data produced by consortia Simons Foundation and MSSNG.
Dani named HHMI Hanna Gray fellow!
https://www.hhmi.org/news/hhmi-names-2024-hanna-gray-fellows
Congrats to former Dennis lab grad student and postdoc Daniela Soto for being named a 2024 HHMI Hanna Gray fellow. She is currently pursuing research at UC Santa Barbara co-mentored by Drs. Soojin Yi and Kirk Lohmueller (UCLA). We are so happy for you Dani!