We are excited to share our new paper published in Autism Research led by former postdoc Sierra Nishizaki, with support from many current (Nick, Gabriana, Gulhan) and former lab members (Tasha, José). Gabriana put together a great summary here.
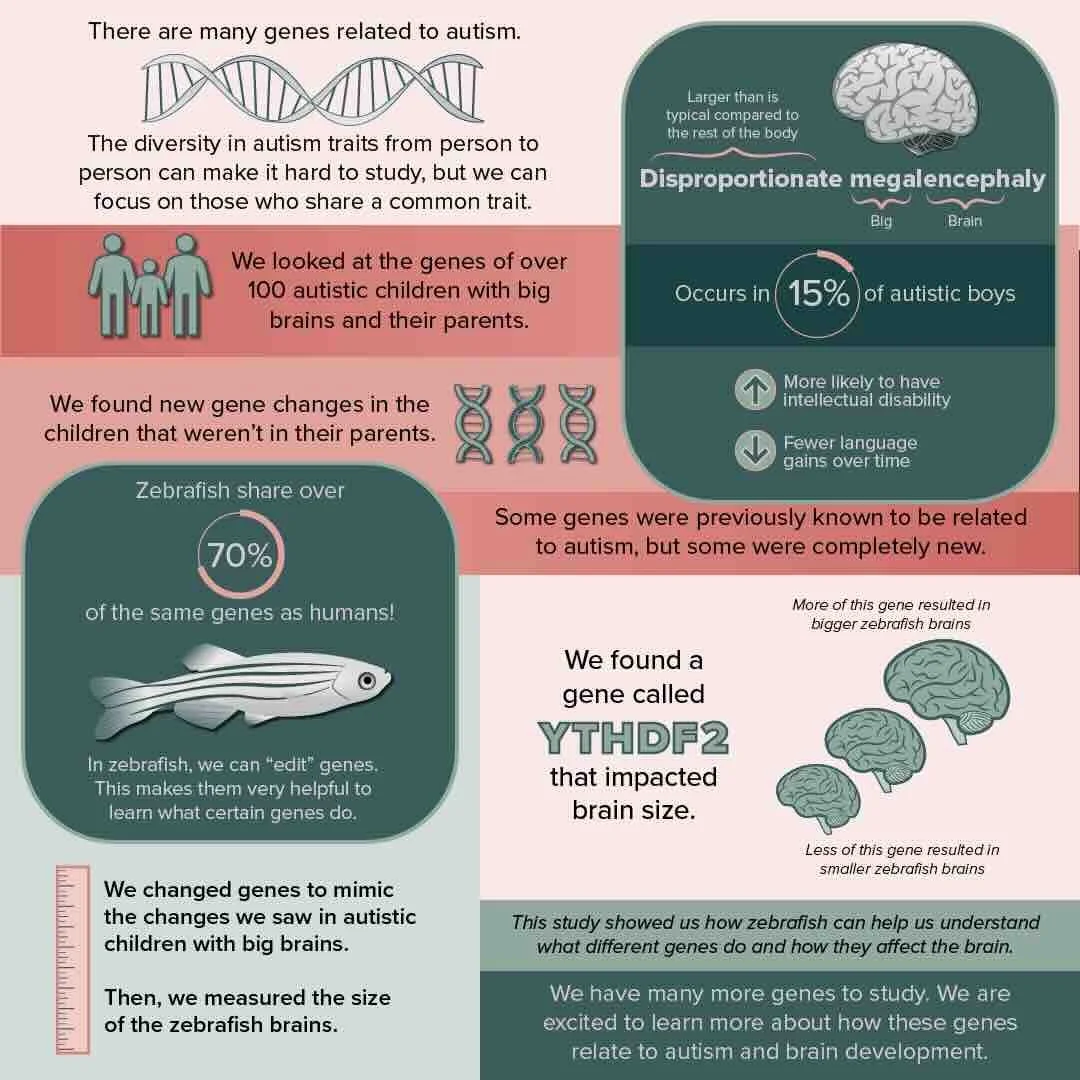
In the study, we identified 154 candidate genes with likely gene-disrupting de novo or rare variants from whole-genome sequences of autistic children with disproportionate megalencephaly. We validated a patient duplication (gain of function) of m6A-mRNA reader YTHDF2 results in enlarged brain in a zebrafish larval model, while knockdown with CRISPR leads to microcephaly. Using single-cell RNA-seq of zebrafish larval brains points to possible downregulation of FMRP-associated genes with increased presence of YTHDF2. We also identified a loss-of-function variant impacting YTHDC1, implicating more broadly the m6A-modification pathway as possibly contributing to ASD.
The project would not have been possible without excellent collaborations through the UC Davis MIND Institute, including the Autism Phenome Project led by Drs. David Amaral and Christine Wu Nordahl, and genomic sequencing data produced by consortia Simons Foundation and MSSNG.