Our recent paper “Evaluation of CRISPR gene-editing tools in zebrafish” led by grad student José Uribe-Salazar is out in BMC Genomics. Several others from the lab contributed including lab manager Gulhan Kaya, volunteer Aadithya Sekar, and postbacs/undergrads KaeChandra Weyenberg and Cole Ingamells.
Some highlights:
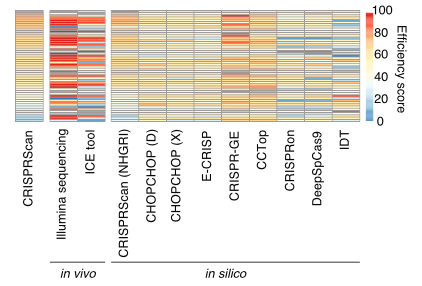
We tested editing accuracy and predictability of 50 guide RNAs (from IDT) with SpCas9 using in vivo, in silico, and in vitro methods.
Large discrepancies existed between in silico editing predictive tools and empirical in vivo cutting efficiencies. The most accurate prediction resulted from using CRISPRScan against a reference specific to our zebrafish strain.
We observed low frequency of off-target mutations with no method (in silico or in vitro) effectively predicting these. More analyses are needed to understand the variability involved in off-target cutting events.
To characterize commonly-used controls in zebrafish CRISPR experiments, we evaluated larvae solely injected with SpCas9 (enzyme or mRNA) and found no evidence of an excess of somatic mutations in these injected larvae.
Performing RNA-seq of injection controls revealed notable variability in gene expression profiles compared to uninjected controls, including genes involved in several metabolic pathways that highlight a potential lasting effect of the microinjection. We recommend caution if performing characterizations of these particular genes in mosaic crispants.
Overall, our hope is for this resource to be helpful for the zebrafish/CRISPR community while they plan their future awesome experiments!