Thank you(!) to the NIH National Institute of Mental Health for funding our lab’s first R01 related to human duplicated genes and their roles in neurodevelopment and autism. We are super grateful and excited to continue work in this exciting research area! We are currently recruiting postdocs to contribute to projects.
Lake Berryessa SUP and kayak adventure!
















Goodbye and hello!



We have much to celebrate!
Undergraduate Gabriana La graduated with a B.S. in Biochemistry and will continue as a PhD student in the UC Davis Integrative Genetics & Genomics program. We are excited for her to return to the Dennis lab to pursue research questions related to autism genetics, funded via an NINDS Diversity supplement.
Undergraduate Jeffrey Zang graduated with a B.S. in Computer Science and will continue as a postbac in our lab until December, where he will move onto greener pastures in Seattle working for Amazon. Congrats Jeffrey!
Recent PhDs Dr. Daniela Soto and Dr. José Uribe-Salazar were the first Dennis lab graduate students to officially walk during UC Davis commencement! Daniela will continue as a postdoc in Dr. Jonathan Flint’s lab at UCLA in October. José will continue as a postdoc with our group to complete final projects (and transfer his vast knowledge to the rest of us!), with plans to transition to biotech in the coming year.
We also are excited to welcome undergraduate researcher, Emily Xu. She will join the group as a UC Davis CIRM COMPASS fellow focused on using zebrafish to model autism candidate genes.
Welcome to Nick, our newest PhD student and T32 MCB trainee
We are excited to welcome our newest Integrative and Genomics PhD graduate student Nick Haghani! Graduating from UC Santa Barbara and taking a postbac year at UC San Diego, Nick’s dissertation project will use zebrafish to test genes implicated in brain development (evolution and conditions). Nick was also selected as a UC Davis MCB NIH T32 trainee! Yayyy!!
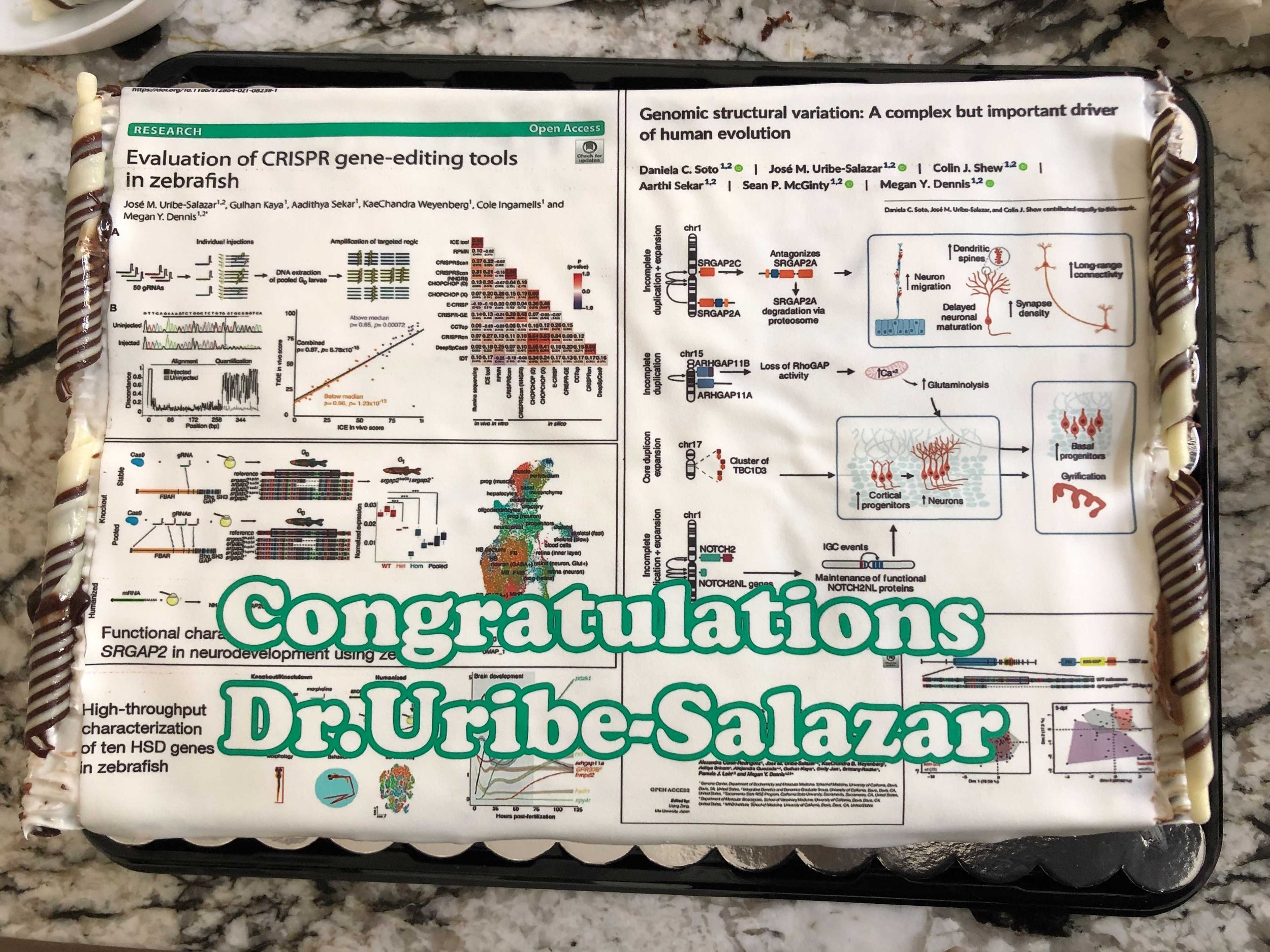
Congrats Dr. José Uribe-Salazar!
We are celebrating Integrative Genetics and Genomics PhD student José Uribe-Salazar for completing his dissertation “Functional screening of human-specific duplicated genes in neurodevelopment using zebrafish“.







Genomic structural variation: A complex but important driver of human evolution
Check out our recently published review in Yearbook American Journal of Biological Anthropology, led by multiple former and current PhD students Daniela Soto, José Uribe-Salazar, Colin Shew, Sean McGinty, and Aarthi Sekar. We share challenges in detecting structural variation and the roles they play in human evolution. In particular, we highlight top candidate human duplicated genes implicated in brain development, detailed below.
Colin heads to Genentech
Recently graduated PhD student Colin Shew will start as a postdoc at Genentech in Dr. Benito-Gutiérrez’s group this spring.
Lab winter outing: Placerville hiking and cider
Red Shack trail
Congrats to Dr. Colin Shew & Dr. Daniela Soto: Our first Dennis lab PhD Grads!
Grad student José Uribe-Salazar presenting at the UC Davis Human Genomics Symposium
Undergrad Gabriana La presenting her very first poster at ABRCMS
Halloween Genome Center symposium
The lab took part in the UC Davis Genome Center Halloween Symposium (first time in-person since 2019). Our Bob-Ross-themed costumes secured our place as team and individual costume winners this year (was stiff competition, as usual!), as well as 3rd place for our Neanderthal-themed pumpkins. Was nice to see faculty, staff, and trainees in person again to discuss topics related to diversity!





Dennis lab at (in-person) ASHG!
We were excited to attend ASHG in-person in LA, to catch up with old friends and make new ones. Four lab members presented their work:
Postdoc Sierra Nishizaki platform talk: “Novel genes associated with the severe autism subphenotype disproportionate megalencephaly”
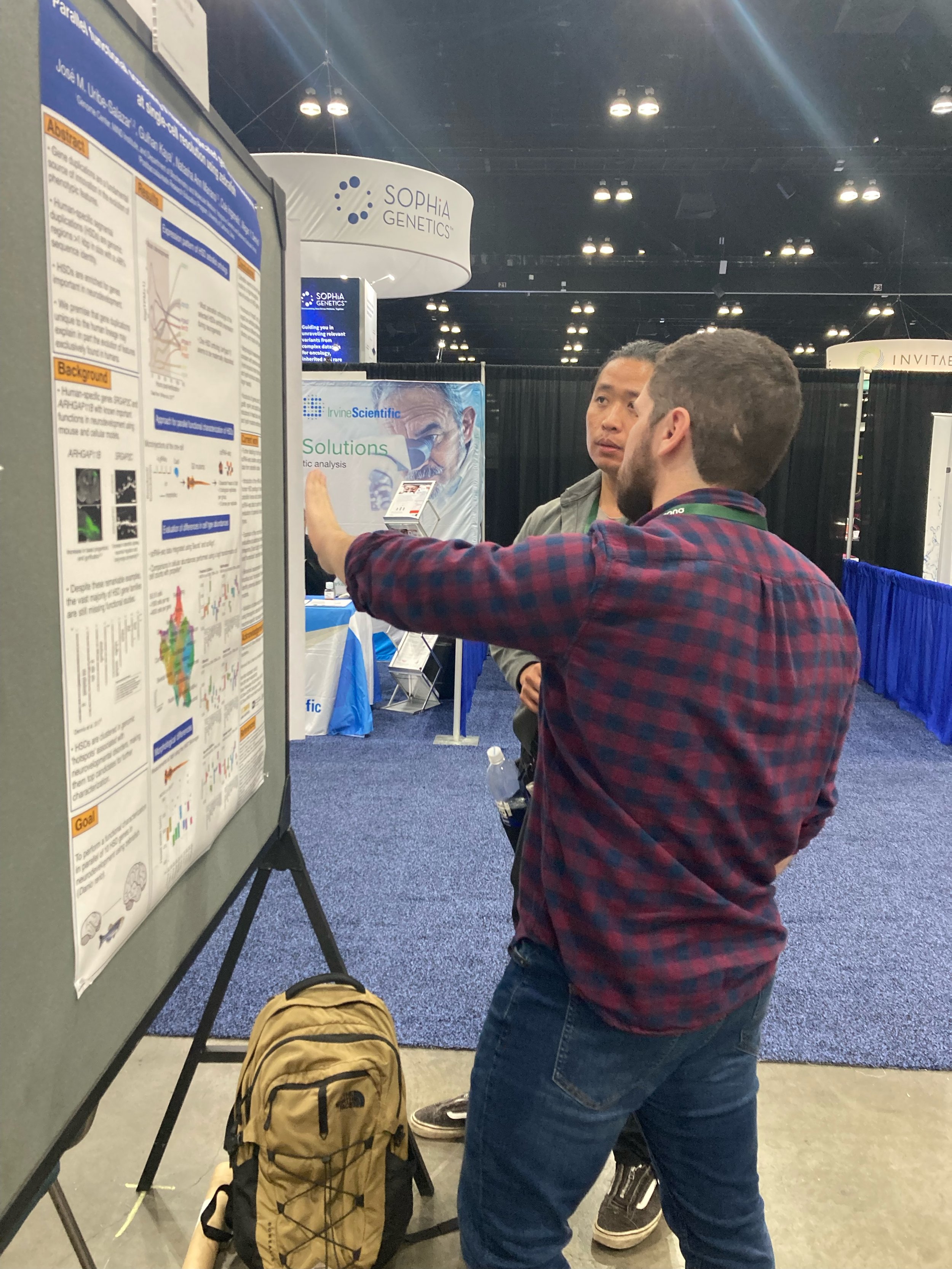
Grad student José Uribe-Salazar poster: “Parallel functional screening of human duplicated genes in neurodevelopment at single-cell resolution using zebrafish”
Grad student Colin Shew poster: “Dissecting mechanisms underlying expression divergence of human duplicated genes”
Grad student Daniela Soto poster: “Population diversity and selection of recent gene duplications detected using a complete human genome sequence”






Good news: Graduations and grad school!
Three junior trainees from the lab are heading to grad school starting in Fall 2022:
Genetics and genomics undergrad Cole Ingamells will be starting the M.S. Bioinformatics program at UC Berkeley.
UC Davis PREP scholar Tasha Mariano is starting in the Ph.D. Molecular Biology and Genetics program at Cornell University.
Former undergrad Aditya Sriram completed his M.S. in Epidemiology from the University of Washington and will be starting in the Ph.D. Human Genetics program at University of Pittsburgh.
We also have farewells and graduations to celebrate:
IGG student Aedric Lim completed his M.S. on “3D Genome Organization in the Developing Macaque Brain” with future plans to pursue a career in genetic counseling. He is currently seeking out clinical research opportunities. Feel free to reach out to Aedric.
Postdoc Sierra Nishizaki completed their ARTP fellowship focused on the genetics of ASD with megalencephaly (with a preprint on the way, stay tuned!) and is currently on the job market as a Bioinformatician (see tweet for more info and feel free to reach out to Sierra).
Former undergrad and postbac KaeChandra Weyenberg completed her M.S. in Public Health from East Tennessee University and is now lead supervisor epidemiologist at the Tennessee Department of Health.
Former undergrad Dhriti Jagannathan completed her M.S. in Genetic Counseling at the University of Minnesota and is now Program Manager and General/Pediatrics Genetic Counselor at Columbia University working with the All Of Us study and eMERGE network.
Dennis lab welcome two new undergrad researchers
Aidan Baraban
A neuroscience major, Aidan will contribute to a collaborative project with the Henn Lab to characterize genes/variants implicated in skin pigmentation variation identified across individuals of South African ancestry using zebrafish.
A day at Rodeo Beach






The complete sequence of a human genome!
A historical scientific milestone has been achieved: the first complete sequence of a human genome (!) published in a special issue of Science. Scientists of the Telomere-to-Telomere (T2T) Consortium sequenced a fully homozygous human cell line, CHM13, generated by the erroneous fertilization of an enucleated egg, to reconstruct the complete genome. The new assembly, named T2T-CHM13, includes 8% of heterochromatin completely missing from the previous human genome assembly (GRCh38). Besides the original Human Genome Project, this is the second most significant progress in our sequencing and understanding of the human genome.
Summary abstract from “A complete reference genome improves analysis of human genetic variation”
Dennis lab members, including Megan and graduate students Colin Shew and Daniela Soto, contributed to a number of the published studies, including:
“A complete reference genome improves analysis of human genetic variation,” where the impact of the complete genome in the analysis of human genetic variation was thoroughly assessed. This was a truly collaborative effort led by four labs (Schatz, McCoy, Zook, and ours). In particular, Daniela performed genome-wide analysis of collapsed duplication in GRCh38, finding 8 Mbp of sequence incorrectly represented in GRCh38 and fixed in T2T-CHM13. Further, we helped demonstrate significant improvements in variant calling across medically-relevant genes.
“Complete genomic and epigenetic maps of human centromeres” by generating CHM13 RNA-seq data, with Colin analyzing transcript abundance across the T2T-CHM13 transcriptome and showing expression of numerous genes near centromeres.
“The complete sequence of a human genome”, with Megan performing comparative genomic analysis of the expanded FRG1 primate-specific gene expansion, which has several missing copies in GRCh38 that are resolved in T2T-CHM13.
This new genome opens many possibilities for scientists who can now explore the most repetitive sequence of the human genome, such as centromeres. Particularly interesting for us is the full resolution of segmental duplications, large historical duplicated segments that are a hallmark of great ape evolution and harbor genes associated with neurodevelopment. Moving forward, we are fully embracing T2T-CHM13 to answer questions regarding the evolution and disease implication of human-specific duplicated genes at unprecedented resolution (and hope many of you will too!).
Additional resources available for the new T2T-CHM13 genome include:
CHM13 open-access data available via GitHub
Dennis lab analysis available via GitHub
Genome stratifications of reference artefacts (incl. collapsed dups) are here
You can read Megan’s commentary about the implications of this scientific achievement in a perspective published in Genome Research. Also, our work, in addition to contributions by Prof Chuck Langley, were highlighted by UC Davis here.
Welcome new lab members!
This past year, we have welcomed two new undergrads, Jeffrey Zang (Computer Science) and Gabriana La (Biochemistry), and a new grad student, Sean McGinty (Integrative Genetics & Genomics).
Gabriana was recently awarded the UC Davis Genome Center Charles and Nanci Cooper Undergraduate Award. Sean received a UC Davis Dean’s Distinguished Graduate Fellowship.
Welcome!!

























